Hydgrogen LDA with Plotted Potentials¶
[1]:
import numpy as np
from CADMium import Psgrid
from CADMium import Kohnsham
#Distance of the nucley from grid center
a = 1.0
#Nuclear charges on centers AB
Za = 1
Zb = 0
#Set polaization. 1 Unpolarized, 2 Polarized
pol = 1
Nmo = [[1]]
N = [[1]]
optKS = {
"interaction_type" : "dft",
"SYM" : False,
"FRACTIONAL" : True,
}
#Grid Options
NP = 7 #Number of points per block
NM = [4,4] #Number of blocks [angular, radial]
L = np.arccosh(15./a) #Maximum radial coordinate value
loc = np.array(range(-4,5)) #Non inclusive on upper bound
#Create and initialize grid object
grid = Psgrid(NP, NM, a, L, loc)
grid.initialize()
#Kohn Sham object
KS = Kohnsham(grid, Za, Zb, pol, Nmo, N, optKS)
KS.scf()
print(f" Total Energy: {KS.E.E}")
iter Total Energy HOMO Eigenvalue Res
-----------------------------------------------------------
1 -0.48889 -2.11156e-01 +1.00000e+00
2 -0.46283 -2.24647e-01 +5.63131e-02
3 -0.45241 -2.29956e-01 +2.30263e-02
4 -0.44831 -2.32059e-01 +9.15620e-03
5 -0.44670 -2.32896e-01 +3.60898e-03
6 -0.44607 -2.33231e-01 +1.41144e-03
7 -0.44582 -2.33365e-01 +5.50624e-04
8 -0.44573 -2.33419e-01 +2.13114e-04
9 -0.44569 -2.33441e-01 +8.16976e-05
10 -0.44567 -2.33450e-01 +3.09185e-05
11 -0.44567 -2.33453e-01 +1.14897e-05
12 -0.44567 -2.33455e-01 +4.15571e-06
Total Energy: -0.4456678603283697
[2]:
#Visualize components
import matplotlib.pyplot as plt
vext,x,y = grid.plotter(KS.vext)
vh,_,_ = grid.plotter(KS.V.vh)
vxc,_,_ = grid.plotter(KS.V.vx + KS.V.vc)
[3]:
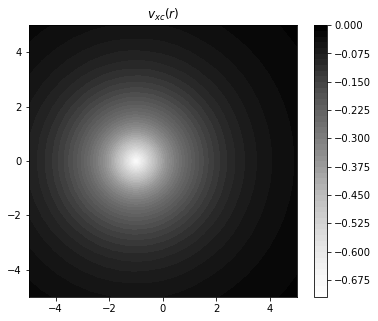
fig = plt.figure(figsize=(6,5))
plt.xlim(-5,5)
plt.ylim(-5,5)
plt.title("$v_{xc}(r)$")
plt.contourf(x,y,vxc,
levels=50,
cmap="Greys",
antialiased = False,
linestyles = "dotted",
)
plt.colorbar()
plt.show()
[4]:
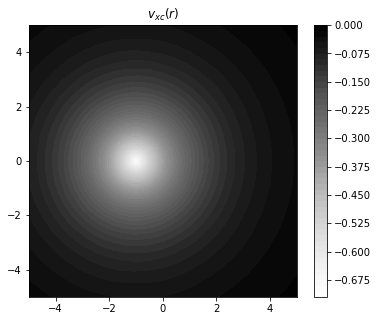
fig = plt.figure(figsize=(6,5))
plt.xlim(-5,5)
plt.ylim(-5,5)
plt.title("$v_{xc}(r)$")
plt.contourf(x,y,vxc,
levels=50,
cmap="Greys",
antialiased = False,
linestyles = "dotted",
)
plt.colorbar()
plt.show()
[5]:
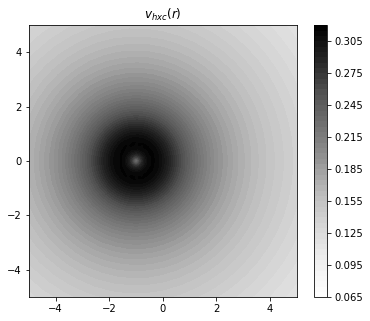
fig = plt.figure(figsize=(6,5))
plt.xlim(-5,5)
plt.ylim(-5,5)
plt.title("$v_{hxc}(r)$")
plt.contourf(x,y,vxc + vh,
levels=50,
cmap="Greys",
antialiased = False,
linestyles = "dotted",
)
plt.colorbar()
plt.show()
[6]:
#Extract components along the z axis
x, v_hartree = grid.axis_plot(KS.V.vh)
_, v_xc = grid.axis_plot(KS.V.vx + KS.V.vc)
[7]:
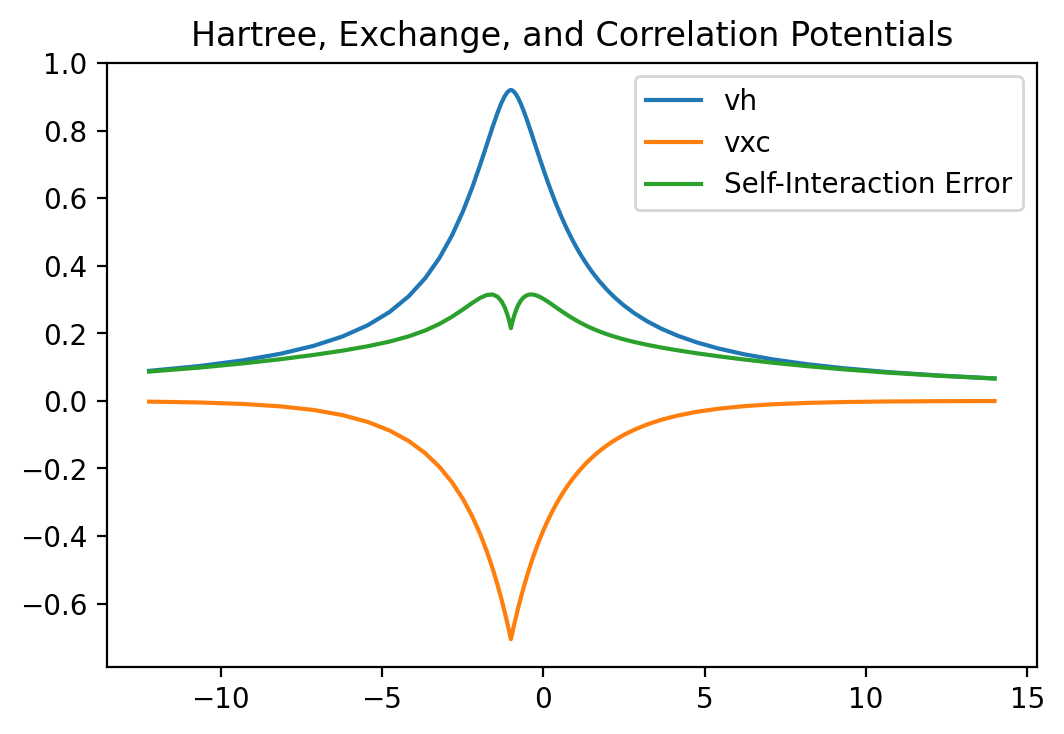
fig = plt.figure(dpi=200)
plt.title("Hartree, Exchange, and Correlation Potentials")
plt.plot(x, v_hartree, label="vh")
plt.plot(x, v_xc, label="vxc")
plt.plot(x, v_hartree + v_xc, label="Self-Interaction Error")
plt.legend()
plt.show()