Stretched PDFT H_2+¶
[2]:
import numpy as np
import matplotlib.pyplot as plt
from CADMium import Pssolver, Psgrid, Partition, Inverter
import CADMium
[4]:
# dis_eq = np.linspace(1.0,5,30)
# dis_st = np.linspace(5.1,10,10)
# dis_eq = np.linspace(1.0,5,10)
# dis_st = np.linspace(5.1,10,3)
# distances = np.concatenate((dis_eq, dis_st))
distances = [1.0,1.5,2.0,3.0,6.0,10]
# distances = [2.0]
energy = []
for d in distances:
a = d/2
Za, Zb = 1,1
pol = 2
#Set up grid
NP = 7
NM = [6,6]
L = np.arccosh(10/a)
loc = np.array(range(-4,5)) #Stencil outline
grid = Psgrid(NP, NM, a, L, loc)
grid.initialize()
#Fragment a electrons [alpha, beta]
#Fragment a electrons [alpha, beta]
Nmo_a = [[1,0]]; Nmo_A = [[1,0]] #Number of molecular orbitals to calculate
N_a = [[1,0]]; N_A = [[0,0]]
nu_a = 0.5
#Fragment b electrons
Nmo_b = [[1,0]]; Nmo_B = [[1,0]]
N_b = [[1,0]]; N_B = [[0,0]]
nu_b = 0.5
#Molecular elctron configuration
Nmo_m = [[1,0]]
N_m = [[1,0]]
part = Partition(grid, Za, Zb, pol, [Nmo_a, Nmo_A], [N_a, N_A], nu_a,
[Nmo_b, Nmo_B], [N_b, N_B], nu_b, { "AB_SYM" : True,
"interaction_type" : "dft",
"kinetic_part_type" : "libxcke",
"hxc_part_type" : "overlap_hxc",
"k_family" : "gga",
"ke_func_id" : 500,
})
#Setup inverter object
mol_solver = Pssolver(grid, Nmo_m, N_m)
part.inverter = Inverter(grid, mol_solver, { "AB_SYM" : True,
"use_iterative" : False,
"invert_type" : "wuyang",
"DISP" : False,
})
part.optPartition.isolated = True
part.scf({"disp" : False,
"alpha" : [0.6],
"e_tol" : 1e-6})
part.optPartition.isolated = False
part.scf({"disp" : False,
"alpha" : [0.6],
"max_iter" : 20,
"e_tol" : 1e-6,
"iterative" : False,
"continuing" : True})
energy.append(part.E.E)
print(f"Done with {d}")
energy = np.array(energy)
# np.save('h2plus_distance.npy', distances)
# np.save('h2plus_overlap.npy', energy)
----> Begin SCF calculation for *Isolated* Fragments
Total Energy (a.u.)
__________________
Iteration A B res
_______________________________________________________
1 -0.24679 -0.24679 1.000e+00
2 -0.24169 -0.24169 1.976e-02
3 -0.24007 -0.24007 6.216e-03
4 -0.23957 -0.23957 1.924e-03
5 -0.23942 -0.23942 5.847e-04
6 -0.23937 -0.23937 1.739e-04
7 -0.23936 -0.23936 4.979e-05
8 -0.23935 -0.23935 1.338e-05
9 -0.23935 -0.23935 3.175e-06
10 -0.23935 -0.23935 5.383e-07
Done with 1.0
----> Begin SCF calculation for *Isolated* Fragments
Total Energy (a.u.)
__________________
Iteration A B res
_______________________________________________________
1 -0.24679 -0.24679 1.000e+00
2 -0.24169 -0.24169 5.569e-02
3 -0.24007 -0.24007 1.732e-02
4 -0.23957 -0.23957 5.343e-03
5 -0.23942 -0.23942 1.624e-03
6 -0.23937 -0.23937 4.820e-04
7 -0.23936 -0.23936 1.380e-04
8 -0.23935 -0.23935 3.708e-05
9 -0.23935 -0.23935 8.799e-06
10 -0.23935 -0.23935 1.492e-06
11 -0.23935 -0.23935 9.391e-08
Done with 1.5
----> Begin SCF calculation for *Isolated* Fragments
Total Energy (a.u.)
__________________
Iteration A B res
_______________________________________________________
1 -0.24679 -0.24679 1.000e+00
2 -0.24168 -0.24168 6.136e-01
3 -0.24007 -0.24007 1.627e-01
4 -0.23957 -0.23957 4.802e-02
5 -0.23942 -0.23942 1.441e-02
6 -0.23937 -0.23937 4.263e-03
7 -0.23936 -0.23936 1.219e-03
8 -0.23935 -0.23935 3.274e-04
9 -0.23935 -0.23935 7.771e-05
10 -0.23935 -0.23935 1.319e-05
11 -0.23935 -0.23935 8.205e-07
Done with 2.0
----> Begin SCF calculation for *Isolated* Fragments
Total Energy (a.u.)
__________________
Iteration A B res
_______________________________________________________
1 -0.24678 -0.24678 1.000e+00
2 -0.24168 -0.24168 6.796e-02
3 -0.24007 -0.24007 2.199e-02
4 -0.23957 -0.23957 6.870e-03
5 -0.23941 -0.23941 2.097e-03
6 -0.23937 -0.23937 6.235e-04
7 -0.23936 -0.23936 1.787e-04
8 -0.23935 -0.23935 4.807e-05
9 -0.23935 -0.23935 1.143e-05
10 -0.23935 -0.23935 1.950e-06
11 -0.23935 -0.23935 1.131e-07
Done with 3.0
----> Begin SCF calculation for *Isolated* Fragments
Total Energy (a.u.)
__________________
Iteration A B res
_______________________________________________________
1 -0.24671 -0.24671 1.000e+00
2 -0.24165 -0.24165 3.192e-02
3 -0.24005 -0.24005 1.024e-02
4 -0.23955 -0.23955 3.196e-03
5 -0.23940 -0.23940 9.781e-04
6 -0.23935 -0.23935 2.919e-04
7 -0.23934 -0.23934 8.410e-05
8 -0.23933 -0.23933 2.280e-05
9 -0.23933 -0.23933 5.502e-06
10 -0.23933 -0.23933 9.806e-07
Done with 6.0
----> Begin SCF calculation for *Isolated* Fragments
Total Energy (a.u.)
__________________
Iteration A B res
_______________________________________________________
1 -0.24595 -0.24595 1.000e+00
2 -0.24120 -0.24120 2.486e-02
3 -0.23966 -0.23966 8.101e-03
4 -0.23918 -0.23918 2.582e-03
5 -0.23902 -0.23902 8.080e-04
6 -0.23898 -0.23898 2.483e-04
7 -0.23896 -0.23896 7.412e-05
8 -0.23896 -0.23896 2.114e-05
9 -0.23896 -0.23896 5.572e-06
10 -0.23896 -0.23896 1.242e-06
11 -0.23896 -0.23896 1.562e-07
Done with 10
[27]:
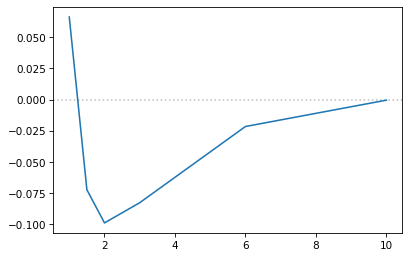
h_energy = -0.24
energy = np.array(energy)
fig, ax = plt.subplots(1,1, dpi=75)
ax.axhline(y=0, alpha=0.5, c="grey", ls=":")
ax.plot(distances, energy - 2 * h_energy)
# ax.set_ylim(-0.12,0.1)
[27]:
[<matplotlib.lines.Line2D at 0x7f7d29a26fa0>]
[14]:
# TEST
[15]:
a = 2/2
Za, Zb = 1,1
pol = 2
#Fragment a electrons [alpha, beta]
Nmo_a = [[1,0]]; Nmo_A = [[1,0]] #Number of molecular orbitals to calculate
N_a = [[1,0]]; N_A = [[0,0]]
nu_a = 0.5
#Fragment b electrons
Nmo_b = [[1,0]]; Nmo_B = [[1,0]]
N_b = [[1,0]]; N_B = [[0,0]]
nu_b = 0.5
#Molecular elctron configuration
Nmo_m = [[1,0]]
N_m = [[1,0]]
#Set up grid
NP = 7
NM = [4,4]
L = np.arccosh(10/a)
loc = np.array(range(-4,5)) #Stencil outline
grid = Psgrid(NP, NM, a, L, loc)
grid.initialize()
part = Partition(grid, Za, Zb, pol, np.concatenate((Nmo_a, Nmo_A)), np.concatenate((N_a, N_A)), nu_a,
np.concatenate((Nmo_b, Nmo_B)), np.concatenate((N_b, N_B)), nu_b, { "AB_SYM" : True,
"interaction_type" : "dft",
"kinetic_part_type" : "libxcke",
"hxc_part_type" : "overlap_hxc",
"k_family" : "gga",
"ke_func_id" : 500,
})
#Setup inverter object
mol_solver = Pssolver(grid, Nmo_m, N_m)
part.inverter = Inverter(grid, mol_solver, {"AB_SYM" : True,
"ENS_SPIN_SYM" : False,
"use_iterative" : False,
"invert_type" : "wuyang",
"disp" : True,
})
#Isolated Fragments
part.optPartition.isolated = True
part.scf({"disp" : True,
"alpha" : [0.6],
"e_tol" : 1e-8})
#Interacting fragments under vp
part.optPartition.isolated = False
part.scf({"disp" : True,
"alpha" : [0.6],
"max_iter" : 200,
"e_tol" : 1e-5,
"iterative" : False,
"continuing" : True})
Total Energy
__________________
Iteration A B res
_______________________________________________________
1 -0.24679 -0.24679 1.000e+00
2 -0.24169 -0.24169 6.139e-01
3 -0.24007 -0.24007 1.627e-01
4 -0.23957 -0.23957 4.803e-02
5 -0.23942 -0.23942 1.441e-02
6 -0.23937 -0.23937 4.263e-03
7 -0.23936 -0.23936 1.218e-03
8 -0.23935 -0.23935 3.271e-04
9 -0.23935 -0.23935 7.760e-05
10 -0.23935 -0.23935 1.314e-05
11 -0.23935 -0.23935 8.375e-07
12 -0.23935 -0.23935 2.442e-06
13 -0.23935 -0.23935 1.718e-06
14 -0.23935 -0.23935 9.537e-07
15 -0.23935 -0.23935 4.791e-07
16 -0.23935 -0.23935 2.283e-07
17 -0.23935 -0.23935 1.053e-07
18 -0.23935 -0.23935 4.763e-08
19 -0.23935 -0.23935 2.124e-08
20 -0.23935 -0.23935 9.382e-09
Total Energy
__________________
Iteration A B res
_______________________________________________________
1 -0.18430 -0.18430 1.000e+00
2 -0.20024 -0.20024 1.459e-01
3 -0.20784 -0.20784 5.906e-02
4 -0.21080 -0.21080 2.194e-02
5 -0.21192 -0.21192 8.064e-03
6 -0.21235 -0.21235 2.963e-03
7 -0.21251 -0.21251 1.089e-03
8 -0.21258 -0.21258 4.001e-04
9 -0.21260 -0.21260 1.470e-04
10 -0.21261 -0.21261 5.400e-05
11 -0.21261 -0.21261 1.984e-05
12 -0.21261 -0.21261 7.296e-06
[34]:
vars(part.E)
[34]:
{'Ea': -0.21261391666219603,
'Eb': -0.21261391666219603,
'Ef': -0.42522783332439207,
'Tsf': 0.6419012008412587,
'Eksf': array([[-0.79224848, 0. ]]),
'Enucf': -1.0955758128947057,
'Exf': -0.3090971334277098,
'Ecf': -0.023736206995989655,
'Ehf': 0.36128011915275443,
'Vhxcf': 0.28330575478424713,
'Ep': -0.6540096116762366,
'Ep_pot': -0.5689834260754593,
'Ep_kin': -0.07871945379408951,
'Ep_hxc': -0.006306731806687867,
'Et': -1.0792374450006288,
'Vnn': 0.5,
'E': -0.5792374450006288,
'evals_a': array([], dtype=float64),
'evals_b': array([], dtype=float64),
'S': 0.36349421310139707,
'F': 0.6961051543833872,
'Ehcor': 0.0,
'Ep_h': -0.03942091105271872,
'Ep_x': 0.02948195670208792,
'Ep_c': 0.0008789269007918529}
[ ]: